Pubnet Instructions
Overview
What is PubNet?
PubNet is a utility that accepts as input up to two PubMed queries, and returns as output a network graph (in multiple
image formats) based on user-specified node and edge selection properties. Nodes represent data items associated with
publications returned by the queries (such as paper ids, author names, and databank ids), and edges represent instances of
shared properties. PubNet can be used to visualize a variety of relationships, such as the
degree to which two authors collaborate or the MeSH Term relatedness of
publications with PDB ids. The visualization is done
with the aid of aiSee.
Generating a graph
- Type or paste any Entrez-PubMed query into the blue box labeled "Query 1". See below for further details.
- (optional) Type or paste a second query into the yellow textbox.
- Choose a Node type and Edge type from the selection boxes at the bottom.
- Click Submit.
Interpreting the Graph
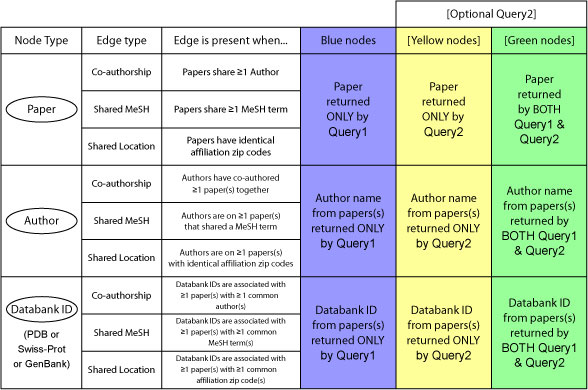
- Graphs are generated by parsing the XML file returned by a PubMed query.
- Each node on the graph represents an entity chosen from Node selection box on the main page.
- Edge is present between two nodes if they share at least one term as chosen from the Edge selection box.
- Edges are colored and (optionally) weighted according to number of terms
(darker & thicker = more shared terms)
- Nodes generated from papers only appearing in the first query set are colored blue.
- Nodes generated from papers only appearing in the second query set are colored yellow.
- Nodes generated from papers appearing in both the first and second query sets are colored green.
Node Selection
- Paper: Each publication (uniquely identified by PMID) returned by the query is represented by a node on the
graph.
- Author: Each author (identified by "FirstInitial LastName") gets a separate node with this option. A single
paper
with several authors will be drawn as several nodes.
- PDB ID: When available, PDB identifiers are included in the XML output of a PubMed query. When this option is
selected, the string "PDB[si]" is appended to the query to ensure only records with PDB ids are returned. Each
PDB id is then represented as a separate node (even if a single paper has multiple PDB ids).
- GenBank ID: Similar to PDB ID, except the string "GENBANK[si]" is used. GenBank ids are then represented as
nodes.
- SWISSPROT: Similar to PDB ID, except the string "SWISSPROT[si]" is used. Swiss-Prot accession numbers are
represented as nodes.
Edge Types
- Co-Authorship: Two nodes are linked by an edge if their respective originating publications have at least one
author in common.
- Shared MeSH Term: two nodes are linked by an edge if their respective originating publications have at
least one MajorTopic MeSH term in common. A MajorTopic MeSH term is defined as having shared term xxxxx, appearing
in the XML output as follows:
<MeshHeading>
<DescriptorName MajorTopicYN="Y">xxxxx</DescriptorName>
</MeshHeading>
or
<MeshHeading>
<DescriptorName MajorTopicYN="N" />
<QualifierName MajorTopicYN="Y">xxxxx</QualifierName>
</MeshHeading>
In many cases, MeshHeadings will have several QualifierNames where MajorTopicYN = "Y". In each case, the QualifierName is
appended to the DescriptorName (separated by a space) and each combination is treated as a separate MeSH term.
- Shared Location:
Two nodes are linked if identical 5-digit numerical codes appeared in their publications' respective
<Affiliation> tags. Note that this simple approach really only works for United States addresses at this time.
Zip codes are extracted using the following regular expression:
/\W(\d{5})\W/.
Because US zip codes use a hierarchical convention, we allow precision to be specified to group
locations that share the first 3 or 4 digits of a 5-digit zip code. For example, a 3-digit prefix is
extracted using the regular expression: /\W(\d{3})\d{2}\W/. In the happy event that there
is demand for support of non-US affiliations, a more sophisticated method may be developed in the future.
Tips for Successful Queries
Complexity of PubNet graphs can scale exponentially with the
number of nodes. It has been my experience that graphs with more than
1000 nodes are difficult to interpret, and they can take a very long time
to load, if they load at all. To get better results, try the following:
Download PubNet Code
The code for PubNet may be downloaded here: PubNet Code

